With the madness of CES over and the Chinese New Year holiday coming up, I finally found some time to catch up on some back issues of Science. I came across a beautiful diagram of the metabolic pathways of one of the smallest bacteria, Mycoplasma Pneumoniae. It’s part of an article by Eva Yus et al (Science 326, 1263-1271 (2009)).
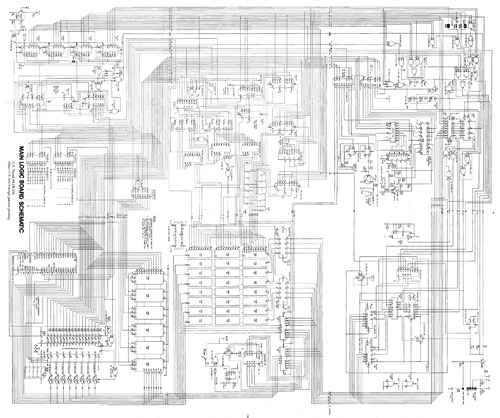
Looking at this metabolic pathway reminds me of when I was less than a decade old, staring at the schematic of an Apple II. Back then, I knew that this fascinatingly complex mass of lines was a map to this machine in front of me, but I didn’t know quite enough to do anything with the map. However, the key was that a map existed, so despite its imposing appearance it represented a hope for fully unraveling such complexities.
The analogy isn’t quite precise, but at a 10,000 foot level the complexity and detail of the two diagrams feels similar. The metabolic schematic is detailed enough for me to trace a path from glucose to ethanol, and the Apple II schematic is detailed enough for me to trace a path from the CPU to the speaker.
And just as a biologist wouldn’t make much of a box with 74LS74 attached to it, an electrical engineer wouldn’t make much of a box with ADH inside it (fwiw, a 74LS74 (datasheet) is a synchronous storage device with two storage elements, and ADH is alcohol deydrogenase, an enzyme coded by gene MPN564 (sequence data) that can turn acetaldehyde into ethanol).
In the supplemental material, the authors of the paper included what reads like a BOM (bill of materials) for M. pneumoniae. Every enzyme (pentagonal boxes in the schematic) is listed in the BOM with its functional description, along with a reference that allows you to find its sequence source code. At the very end is a table of uncharacterized genes — those who do a bit of reverse engineering would be very familiar with such tables of “hmm I sort of know what it should do but I’m not sure yet” parts or function calls.
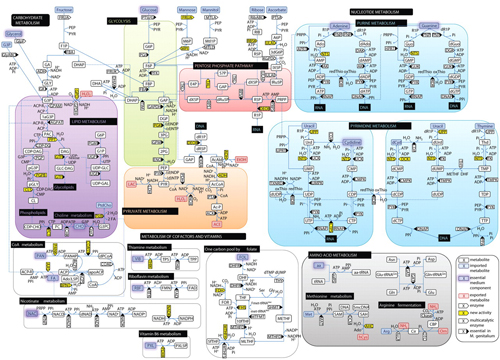
Bacteria (and even that mycoplasma) are a lot more complicated than that diagram shows. The word “metabolism” doesn’t encompass everything the cell does; commonly it only refers to the major energy pathways — although that diagram of “metabolism” includes some other stuff, such as processing of vitamins. Suppose the mycoplasma cell wants to change its shape, for instance. (There is a species of mycoplasma which has a point at one end, which it uses to penetrate into our cells, the better to infect us. It’s not the same species of mycoplasma as in the diagram; but the diagrammed species, too, must have some way or other of controlling its shape: even if it just wants to be a sphere, the radius of that sphere needs to be regulated somehow.) That diagram of metabolism doesn’t show a single part of the chemistry (let alone the physics) of that operation. Likewise, that list of enzymes isn’t a complete BOM for the whole organism, but just the enzymes involved in metabolism. It’s sort of like showing the ALU of an Apple II — an important part, but by no means all.
“That diagram of metabolism doesn’t show a single part of the chemistry (let alone the physics) of that operation.”
Neither does the Apple schematic.
Yes, but in the Apple schematic, the chemistry and physics are well-defined lower layers, whose operations need not be considered when analyzing the schematic. Someone else designed the NAND gates, and the main thing the schematic reader needs to know is that they are NAND gates; a knowledge of solid state physics was necessary to make them but is immaterial when it comes time to use them. In biology, everything is mixed together; there is no layering. Layering is a human contrivance, intended to make things easy for engineers. Evolution knows no such boundaries. Some biological functions depend on the physics of diffusion; others on chemical equlibrium; others on quantum effects; and all these are mixed together within a single cell.
(Of course the really great engineers do know details of lower layers, and occasionally take advantage of them to do something “impossible”. But that is a rare feat, not an everyday practice.)
I vividly remember the moment as a teenager when my understanding of the Apple ][ memory/video/refresh design snapped into place. Like Bunny, I had also been fascinated by the idea that the machine could be described by a diagram. I had spent days reading the TTL data book alongside the schematic. When it all snapped, I had an immense appreciation for Woz’s genius of minimal design.
That moment led to everything that followed – my degree, my interest in reverse engineering, my career in hw and sw design, and (most recently) over ten years of getting paid for open source development.
I still have that Apple ][ technical manual and the print that came with it.
I love reading about and looking at stuff like that. I remember when I was reading about DNA coding and thinking “wow, that’s similar to how data is coded onto magnetic tape!” It’s neat to look at these sorts of visualizations and muse on how the various moving parts interact.
Thought I would share a very cool website about the 6502 processor for those interested. You can simulate a 6502 down to the transistor level in your browser.
http://www.visual6502.org/
I’m not a part of the above site. It was too cool to pass up and not say anything!
I was looking at the Mycoplasma diagram and thinking “wow, there are actually people out there who can make sense of this!”. Then i looked at the Apple II one and thought “Yeah, that makes sense”, although I hadn’t appreciated when I first looked at it years ago that it used unbuffered I/O… how could it handle having a pile of arbitrary cards plugged into it?
Hello. I like this article about A schematic for M. pneumoniae metabolism bunnie's blog. It was fine. I own a blog too, but about eBooks for Kindle for example bossy pants or in the garden of beasts love terror and an American family in Hitlers Berlin. Keep at it
Kindlebook